RNase P – bewildering diversity and evolutionary puzzle
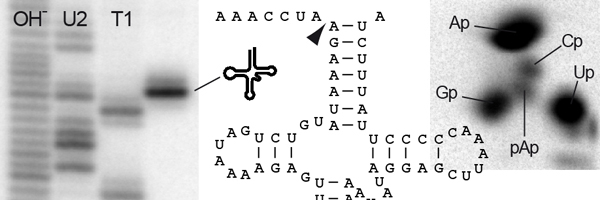
The extensions on the 5' (front) end are clipped off by an enzyme called RNase P [for review see ref. 2]. The RNase P family is remarkable for the heterogeneous makeup of its members. For almost three decades all RNase P enzymes were believed to be ribonucleoproteins, i.e., composed of RNA and protein. Noteworthy, the RNA is the catalytically active moiety of this type of RNase P. This RNA is widely believed to be a remnant from an ancient prebiotic chemistry dominated by RNAs (RNA world) that persisted up to modern forms of life; for unknown reasons RNase P RNAs resisted the general trend towards protein catalysis. Nevertheless, there seems to be a trend towards more and more proteins being recruited to the ribonucleoprotein by these RNAs: whereas bacterial RNase P requires just one small protein in addition to its RNA, 10 proteins are required to complement human nuclear RNase P. The reasons or biological meaning of this increase in complexity are currently not known.
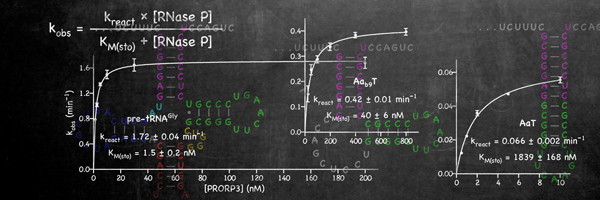
Recently, our lab demonstrated that this story of RNase P as an RNA enzyme is incomplete. First, the identification of the molecular components of human mitochondrial RNase P revealed this enzyme to be composed of three proteins only, and no RNA [3]. Related, yet more simple RNase P enzymes were subsequently found in the mitochondria and chloroplasts of plants and protists [4-6]. This type of RNase P is composed of a single protein, homologous to the nuclease subunit of human mitochondrial RNase P, and we now call these proteins PRORP, for proteinaceous or protein-only RNase P. In the end, their localization turned out not to be restricted to organelles, but PRORPs also act as nuclear RNase P in plants and some protists [5,7]. In summary, our findings essentially changed the paradigm of RNase P as an RNA enzyme, and revealed the unprecedented and bewildering diversity of an enzyme family as the apparent result of convergent evolution at the molecular level.
Evolution apparently found a simple and efficient way to make tRNAs without RNA-based catalysis and it appears thus more astounding than ever that an RNA enzyme has persisted at all for the seemingly simple biological process of tRNA 5'-end formation. Why have RNA enzymes been retained in so many phylogenetic groups, even at the cost of maintaining highly complex, multi-subunit ribonucleoproteins, when apparently a single protein can substitute for the same function in other eukaryal systems? It is one of our aims to elucidate the evolutionary constraints underlying this apparent paradox by an in-depth comparison of proteinaceous RNase P enzymes with RNA-based forms.Human mitochondrial RNase P – a multi-enzyme machine The enzyme found in human mitochondria is probably the most unusual member of the RNase P family. The nuclease subunit is homologous to the proteinaceous enzymes (PRORPs) found in plants and protists [3,6]. However, different from those, human PRORP acts in association with two more proteins, both of which have additional, independent enzymatic roles [ 8]. One (SDR5C1) is a dehydrogenase involved in the degradation of branched-chain amino and fatty acids. Together with the other one (TRMT10C) it forms a methyltransferase complex responsible for the N1-methylation of purines at position 9 of tRNAs. The TRMT10C-SDR5C1 methyltransferase complex in turn acts together with PRORP in the cleavage at the 5’ end of tRNAs. The relationship and interplay of the three different enzymatic activities gathered in the human mitochondrial RNase P complex is poorly understood and another major research interest of our group.
More tRNA maturation enzymes
TRMT10C is one of three related methyltransferases in human cells [8]. The other two, TRMT10A and TRMT10B, localize to the nucleus rather than to mitochondria. All three enzymes methylate guanosine at tRNA position 9, but only the TRMT10C-SDR5C1 complex is able to methylate also adenosine at this position. TRMT10A and TRMT10B do not require a protein partner for activity. Whereas the methylation of mitochondrial tRNAs appears crucial for their correct folding, the role of the same modification in the nuclear-encoded, cytosolic tRNAs is unclear.
The 3’-end processing enzyme RNase Z is shared between the nucleus and mitochondria, i.e., a single gene gives rise to both enzyme isoforms [9]. Like RNase P, mitochondrial RNase Z plays a crucial role in the general processing of the mitochondrial transcriptome (see below).Mitochondria, their tRNAs, transcript processing, and disease Coming soon!
References
- Phizicky, E. M., and A. K. Hopper (2010).
tRNA biology charges to the front. Genes Dev. 24, 1832-60. - Lai, L. B., A. Vioque, L. A. Kirsebom, and V. Gopalan (2010).
Unexpected diversity of RNase P, an ancient tRNA processing enzyme: challenges and prospects. FEBS Lett. 584, 287-96. - Holzmann, J., P. Frank, E. Löffler, K. L. Bennett, C. Gerner, and W. Rossmanith (2008).
RNase P without RNA: identification and functional reconstitution of the human mitochondrial tRNA processing enzyme. Cell 135, 462-74. - Gobert, A., B. Gutmann, A. Taschner, M. Gößringer, J. Holzmann, R. K. Hartmann, W. Rossmanith, and P. Giegé (2010).
A single Arabidopsis organellar protein has RNase P activity. Nat. Struct. Mol. Biol. 17, 740-4. - Taschner, A., C. Weber, A. Buzet, R. K. Hartmann, A. Hartig, and W. Rossmanith (2012).
Nuclear RNase P of Trypanosoma brucei: a single protein in place of the multi-component RNA-protein complex. Cell Rep. 2, 19-25. - Rossmanith, W. (2012).
Of P and Z: Mitochondrial tRNA processing enzymes. Biochim. Biophys. Acta 1819, 1017-26. - Gutmann, B., A. Gobert, and P. Giegé (2012).
PRORP proteins support RNase P activity in both organelles and the nucleus in Arabidopsis. Genes Dev. 26, 1022-7. - Vilardo, E., C. Nachbagauer, A. Buzet, A. Taschner, J. Holzmann, and W. Rossmanith (2012).
A subcomplex of human mitochondrial RNase P is a bifunctional methyltransferase–extensive moonlighting in mitochondrial tRNA biogenesis. Nucleic Acids Res. 40, 11583-93. - Rossmanith, W. (2011).
Localization of human RNase Z isoforms: dual nuclear/mitochondrial targeting of the ELAC2 gene product by alternative translation initiation. PLoS ONE 6, e19152.